Note for: Biomarker-Guided Development of DNA Repair Inhibitors
Note for: Biomarker-Guided Development of DNA Repair Inhibitors
doi: 10.1016/j.molcel.2020.04.035
Key theme;
Drug targeting DNA damage response exploit the synthetic lethality
Targeting the cells which are defective in DNA repair using PARPi
Another selective agents for DNA repair -- >
ATR
CHK1
WEE1
ATM
cNHEJ
Alt EJ
Review biomarkers which aid physician to decide when to use these inhibitors
(in the old day) Oncologist used drugs targeting DDR which has cytotoxic property
Alkylating agent temozolomide -- > repair by MGMT
Cisplatin -- > repair by NER and ssDNA repair
Bleomycin -- > cleave DNA and generating DNA double strand break
Topo-I and II inhibitor -- > causing ssDNA break and dbDNA break, respectively
DSB
V(D)J recombination
High error by cNHEJ -- > diversity of T-cell receptors and antibodies
cNHEJ repair
leading to characteristic DNA damage lesions
large deletion
loss of hetrozygosity
telomeric allelic imbalance
HR deficiency
Genomic scar
Trio of three proteins
MRE11-RAD50-NBS1 (MRN complex)
Recognize DSB
Inhibitor of ATM and RAD51 – currently being developed
PARPi
Targeting BRCA1/2 defective
Backup pathway -- > BER
DSB
HR relies on the availability of 3’ overhang
53BP1+shieldin complex (SHLD1-3, REV7) -- > block further resection by nuclease
Thus, it can promote NHEJ
53BP1 and BRCA1 -- > play opposing role
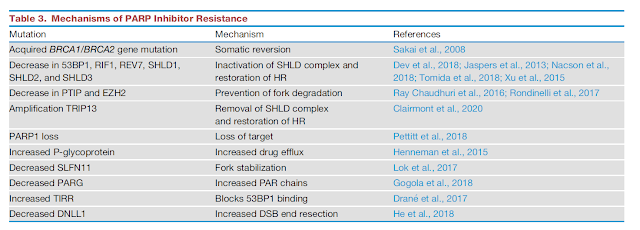
Inhibitor toward TRIP3 (amplified in many BRCA1-deficienct tumor) -- > might be useful to prevent PARPi resistant
PARP resistance
Intrinsic -- > naturally resistant
Extrinsic -- > acquired resistant
Alt-EJ dependent on PARP1
DSB -- > MSN and PARP1 recognizes break (requires 2-20 bp homologous DNA sequence) -- > Pol theta mediates repairing
In HR proficient -- > alt NHEJ rarely occur -- > in HR deficiency -- > tumor seems to relied on alt NHEJ
Thus, Possibly -- > Pol theta is a good target for HR deficiency
Single-strand annealing
Is DSB repair
Not similar to alt-NHEJ
Single strand annealing is inhibited by RAD51 and require RAD52 to recombines homologous ssDNA
Mode of mechanism
Relying on synthetic lethality, ssDNA could not be repaired due to PARP1i -- > causing accumulation of DSB during cell division -- > lack of HR -- > forcing the cell to repair through cNHEJ -- > toxic
Treating with the 3rd gen -- > reveal the toxicity correlates to PARP trapping ability at DNA damage site -- > PARP-DNA complex is lethal in HR defect
Inhibition of Pol-theta dependent altEJ
Pol-theta
Has RAD51 binding site, ATPase, and helicase activity
Pol-theta can remove RAD51 from DNA during repairing process
If RAD51 cannot be removed – it becomes toxic to the cell -- > it disrupt repairing process
Inhibition of Pol-theta -- > good for both
HR-deficient cells by inhibiting alt-NHEJ
cell acquired PARPi resistance via HR restoration
Mechanism of resistance in HR-deficient tumors
efflux of PARPi -- > upregulation of drug-efflux transporters
Mutation of PARP which disrupt DNA binding (like PARP(-/-))
Restoring HR repair
Re-establishing replication fork stability
Observation
Patients who resist to Pt-based therapy -- > also correlate with resist PARPi
But not in the case of cisplatin resistant
Predictive and pharmacodynamic biomarker for DDR inhibitor drug development
Mostly detecting mutation in genes
BRCA1/2
PALB2
RAD51C
RAD51D
Few on methylation state
Signature 3
Genomic mutational signature – signature 3 -- > associated with HR repair deficiency -- > still having some patients showed no clear mutation in BRCA1, BRCA2 or PALB2.
Requirement for WES or WGS -- > identifying by signature 3 (programming to analyze the HR deficiency profile) -- > but it is not widely available
Clinical-grade targeted sequencing panels -- > being developed to detect signature 3
Possibility if acquired resistance to PARPi through genetic and non-genetic mechanism -- > highlight the need for functional biomarkers -- > determine HR proficiency -- > immunofluorescence-based RAD51 assay
RAD51 foci formation
Critical step for HR pathway
Can differentiate between HR-proficient cancers and HR-deficient cancer
Using irradiated live tumor cells -- > predictive of HR-deficient breast cancer
Live cell is normally not available -- > attempt to perform RAD51 assay on formalin fixed samples
Platinum sensitivity – correlate with PARP sensitivity -- > observation from ovarian cancer
Progress on developing integrative genomic assays predict HRD
Myriad genetics -- > HRD assay -- > based characteristic genomic findings -- > LOH, telomeric allelic imbalance (TAI), and large-scale state transition (LST)
Foundation Medicine T5 NGS assay -- > assess mutation status of 30 HR genes -- > calculate percentage of LOH
These two have been used to predict the PARPi (niraparib and rucaparib) sensitivity in large clinical trials, phase 3 -- > ovarian cancer
Fail to identify all platinum-sensitivity patients who who benefited from PARPi
Thus -- > Pt-sensitivity can be used as surrogate marker for PARPi responsiveness in ov ca
Pharmacodynamic assessment
PARP inhibitor + Pol-theta -- > cause more RAD51 foci formation and becoming toxic to the cell
PARP inhibitor + IDH1/2 (mutation) -- > cause more sensitivity to PARP
Two major acquired PARPi resistant
Restoration of HR
Stability at the replication forks
Inhibition of ATR/CHK1/WEE1 pw -- > cause complication -- > myelosuppression -- > might be hard when combining with PARPi
Strategies designed to increase replicative stress
Cancer has inherent degree of endogenous replicative stress
Top I and II inhibitors are thought to be a good strategic approach
Increasing the replicative stress by decreasing dNTP through the use of hydroxyurea and gemcitabine
Combination of PARPi with cytotoxic agents
Topo-I cleavage cpx is stabilized by PARP1
Complication occurs when perform combination; full dose chemotherapy could not be done easily -- > overlapping myelosuppression
Combination could also increase normal tissue becoming toxic
Normal tissue vs cancer tissue -- > given same activity -- > no advantage can be achieved when combining with cytotoxic agent
Cancer cell
Have greater baseline deficiency in DDR
More damage than normal cell in general sense
Using this different to increase therapeutic index
By doing this -- > we can observe the DDR defective profile in cancer
Combination of DDR inhibitors with immunotherapy
There is a successfulness story regarding of MMR -- > more mistakes -- > generate neoantigen -- > good for immunotherapy
Not all defective DNA repair pw can give rise to neoantigen which later provokes good immune response
But, DNA damage can induce the expression of PDL1 -- > increase the amounts of targets which will be recognized by PDL1i
Conclusion
Besides the successfulness of PARP1i on BRCA1/2 in OVCA, Breast and pancreatic cancer
More drugs targeting DNA damage and repair are gradually entering clinical phase
ATM
ATR
CHK1
DNAPK
WEE1
Things to concern when making the combination with DDR
Optimal clinical setting
Dose intensity
Drug scheduleing
Predictive biomarkers to guide appropriate drugs in clinical trial to initiate DDR inhibitors -- > PD biomarkers should be incorporated to reveal whether DDRi precisely hit the target.
More understanding of resistant regarding on DDR alone or combination to prevent acquired mutation or use it for reversing the resistant









Comments
Post a Comment