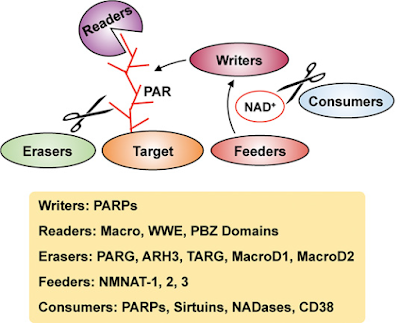
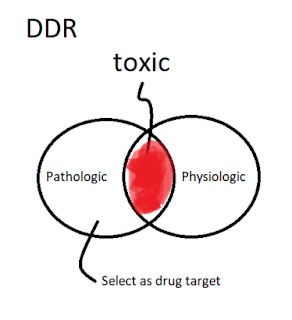
Note for the youtube lecture: Computational Drug Discovery: Machine Learning for Making Sense of Big Data in Drug Discovery
Note for the youtube lecture: Computational Drug Discovery: Machine Learning for Making Sense of Big Data in Drug Discovery Link: https://youtu.be/uoVAd_zd-90 Drug 1. Biological entity -- biologic 2. Chemical based drugs – synthetic drugs, natural product, small molecule Drug discovery – for one particular drug - 10-15 years - Failure rate -- >90% - Cost ~2 billion USD Drug discovery process (million to one compound and it could fail!!!) 1. Identified target; ~30,000 protein (not include PTM processes) 2. Screen for the hit compound – molecule disrupted the activity of particular protein 3. Optimization of the hit compounds – called medicinal chemistry, scaffold hopping, bioisostere, structure-activity relationship – getting the potential compound (potent) 4. ADMET (balance between potency and toxicity) Computational drug discovery - Green chemistry; safe way to generate t