Note for: Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas
Note for: Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas
(doi: 10.1016/j.celrep.2018.03.076)
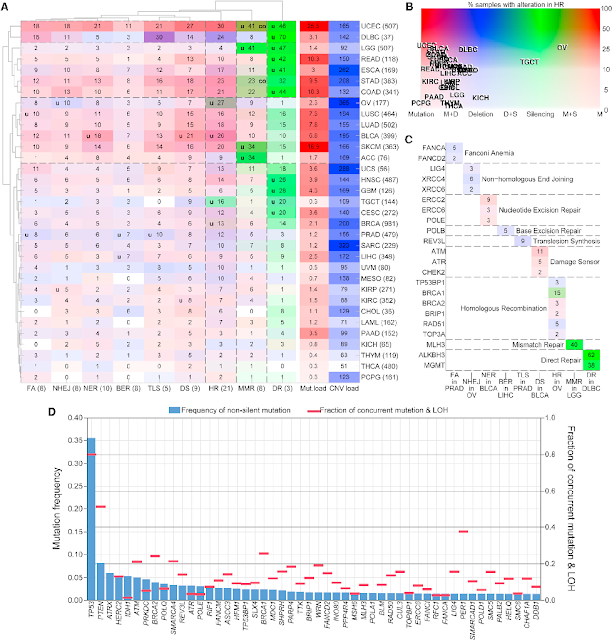
The Cancer Genome Atlas (TCGA) Pan-Cancer analysis of DNA damage repair (DDR) deficiency in cancer. using integrative genomic and molecular analyses the data from cancer (33 cancer), basically focus on DDR. HR and directed repair are the most frequency altered.
We systematically analyzed somatic alterations to provide a comprehensive view of DDR deficiency across 33 cancer types. DDR genes can be grouped into functional pathways defined by genetic, biochemical, and mechanistic criteria.
Type of damages;
1. DNA base damage -BER, NER and DR
2. Mismatch - MMR
3. DNA strand break - HR, NHEJ, FA, and TLS
All of the major DDR pathways, with the exception of the FA pathway, have been identified in virtually all organisms. This reflects the universal need to counter the chemical instability of DNA and repair additional damage. Frequent TP53 somatic mutations in many cancer types can disrupt the DNA damage response, apoptosis, or senescence pathways active in many early-stage cancers. DDR deficiency may also lead to specific mutational ‘‘signatures,’’ e.g., the short tandem repeat instability linked to the inactivation or silencing of DNA MMR in colorectal, ovarian, or endometrial cancer. DNA interstrand crosslinks (ICLs) and double strand breaks may be particularly difficult to repair, requiring coordination of the NER, BER, FA, and HR pathways. The resulting consequences across 33 different human cancer types. The loss of specific DDR pathways in cancer, in contrast to other cellular ‘‘hallmarks’’ of cancer, often generates stable—and thus more readily interpretable—‘‘footprints’’ in cancer genomes, detected as an increased mutation burden, altered mutational signatures, or copy-number alterations including loss of heterozygosity (LOH).
Analysis of 276 genes;
- involved in major DNA repair pws
- 208 is annotated to one or more specific DDR pw.
- the rest 68 genes is annotated to key DDR-related pathways
- core-DDR --> 71 genes for DNA repair pw specific + 9 DNA damage response genes
Prevalent DDR Alterations across Cancer Types;
- determine the mutation burden of cancers
- looking for somatic truncating and missense mutations, deep copy-number deletions, epigenetics silencing
- 9125 PanCanAtlas samples (33 cancer types, 10 pws)
- Cancer types with higher global mutation burden --> higher mutation frequency in DDR pw.
- Cancer types with large number of somatic copy-number alterations (SCNAs) -- having a larger number of SCNAs in DDR pw.
It is already well established that cancers with somatic POLE and POLD1 mutations in their exonuclease domains or with microsatellite instability (MSI) exhibit a substantially higher mutation burden. whether DDR genes would be identified as cancer drivers using mutation frequency-based prediction methods.
Driver prediction tool (computer algorithm, not a wet lab);
1. 20/20+
2. Mut-Sig2CV
3. OncoDriveFML
4. MuSiC2
5. CompositeDriver
Using these tools are able to identified 48 DDR genes as potential drivers.
- 8 putative DDR driver genes were unique to a cancer type
- 18 were identified only as part of a PanCanAtlas analysis
- 23 remaining were identified in analyses of one or more individual cancer type as well as in PanCanAtlas analyses.
Epigenetic silencing was identified as an alternative prominent mechanism leading to recurrent gene deficiency.
Stringent calling criteria;
-identifying 12 DDR genes -->strongly correlated with methylation-driven transcriptional silencing.
Genomic Instability Linked to HR Deficiency and
Prognosis
-6 different SCNA scores --> used to characterize the extent of aneuploidy, LOH, and homologous recombination deficiency
Genomic scarring with large-scale genome instability has been attributed to homologous recombination deficiency (HRD). Alterations in either BRCA1 or RAD51B increased HRD score in most cancer types including the top two cancer types with the greatest number of alterations in either gene.
Gene fusion databases;
1. ChimerDB 3.0
2. TCGA Fusion Gene Data Portal
Potential functional consequences of relatively rare, recurrent mutations in 22 DDR genes by protein structural analyses and modeling. included MGMT, PARP1, TOP3A, BLM, ERCC2, HFM1, POLE, POLD1, and POLQ, which collectively harbored 1,380 unique rare, recurrent non-synonymous mutations.
Thus, molecular modeling and protein dynamics in concert may reveal mechanisms by which somatic mutations alter DDR protein function. We compared both mutation and SCNA burden scores for genes with predicted strongly destabilizing versus non-destabilizing mutations.
Machine-Learning-Derived Expression Signature Predicts TP53 Inactivation;
The loss of TP53 function across many cancer types has significant functional consequences as measured by genomic instability in association with a higher SCNA burden and increased HRD scores. Cancer-associated TP53 mutations may promote these consequences through simple loss of function, as well as by altering transcription or through dominantnegative, gain-of-function mechanisms.
We computed multiple DDR footprint scores based on quantitative estimates of DNA damage and investigated their association with clinical outcomes. We tested DDR footprint score associations with overall survival (OS) and progression-free interval (PFI) across 28 cancer types by fitting Cox proportional hazards models, using survival outcome data. We used TCGA PanCanAtlas data to systematically analyze the prevalence, nature, and consequences of DDR gene and pathway alterations across 9,125 samples representing 33 different cancer types.
which many DDR gene alterations might increase specific mutation types, as well as overall mutational burden. For example, we found a previously undefined signature 8 was strongly associated with BRCA deficiency, especially BRCA1. EXO5 deficiency, identified here as an often epigenetically silenced DDR gene, was associated with signature 1 across multiple cancer types and has been associated with the poor clinical outcomes. For example, a combination of protein structural modeling and molecular dynamics simulations were used to predict the functional consequences of rare, recurrent nonsynonymous mutations in 22 DDR genes. We found that POLD1 mutations, despite being less common than the POLE or POLQ mutations that contribute to hereditary colorectal cancer risk, and the hypermutated phenotype, were as strongly associated with genomic instability. Molecular dynamics simulations further identified a subset of DDR gene mutations with the potential to alter protein conformational changes independent of effects on protein stability, raising the provocative question of whether destabilizing mutations alone contribute to genomic instability in cancer. A second extension of PanCanAtlas genomic data was to use machine learning to predict TP53 inactivation status from gene expression data.
we identified associations among most DDR footprint scores and clinical outcomes after controlling for covariates such as age, cancer grade, and stage. We also identified HRD-high cancers including subsets of ovarian, uterine, lung squamous, esophageal, sarcoma, bladder, lung adenocarcinoma, head and neck, and gastric carcinomas. Virtually all of these subsets of cancers may have enhanced responsiveness to platinum-based compounds that are given as standard-of-care therapies. These results indicate the potential of HRD scoring to predict both platinum response and PARP inhibitor sensitivity.
Thus, combination therapies that induce or potentiate replication stress or impair replication fork protection may be particularly effective in killing HR-deficient cancers. DDR pathway deficiencies are also mechanistically linked to mutation burden and mutational diversity. Thus,DDR pathway deficiencies in cancer may potentiate immune-based therapies by driving neoantigen production to enhance immune recognition and targeting thus may identify subsets of patients with a higher likelihood of responding to immune-based therapies.










Comments
Post a Comment